With rare exceptions, each of the trillions of cells in our bodies carries an exact duplicate of the human genome, which contains between 20,000 and 25,000 protein-coding genes. But to carry out the specialized functions that make life possible, organs like the kidney, lung, heart, and brain rely on tissues built from distinctive cell types, which come about when individual cells develop to express only a particular subset of genes in the genome.
Until recently, the diversity of gene expression across cell types, known as the transcriptome, has been difficult to decipher. But with the rapid rise of single-cell biology, scientists have created tools and techniques that reveal precisely which genes are expressed by the individual cells that make up tissues and organs. In addition to bringing a deeper understanding of normal biology, these single-cell approaches promise to open up avenues to new therapies, because diseases generally strike specific cell types.
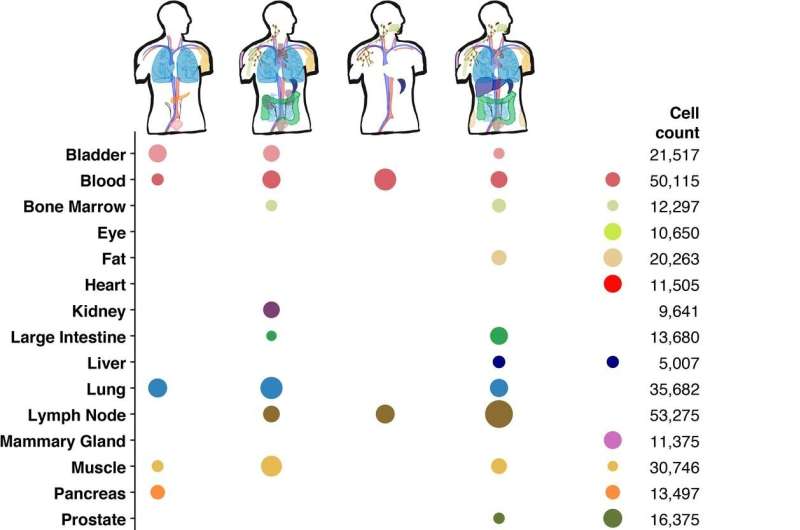
Now, in a paper published in Science, the Tabula Sapiens Consortium, a team of more than 160 experts led by scientists at the Chan Zuckerberg Biohub, has unveiled a massive digital atlas that maps gene expression in nearly 500,000 cells from 24 human tissues and organs, including the lungs, skin, heart, and blood.
The Tabula Sapiens cell atlas is the largest to include multiple tissues from the same human donors, and the first to include histological images of the tissues, and to incorporate details of the microbial communities living alongside the human cells that make up the various compartments of the gut.
“The quality and breadth of these data are unparalleled,” said senior author Stephen Quake, D.Phil., the Lee Otterson Professor of Bioengineering and professor of applied physics at Stanford University, for whom Tabula Sapiens is the fulfillment of a 20-year “obsession.” Quake, who is also president of the CZ Biohub Network, added, “This atlas will allow scientists to ask and answer questions about human health and disease that they’ve never been able to approach before.”
The new paper is one of four major collaborative studies published in Science this week, all of which have created comprehensive and openly available cross-tissue cell atlases as part of the international Human Cell Atlas (HCA) consortium. HCA is supported by a wide range of global funders including the Chan Zuckerberg Initiative (CZI). Additional funding for Tabula Sapiens was provided through CZI’s Single-Cell Biology Program.
“This collection of work is truly inspiring,” said Jonah Cool, Ph.D., science program officer for single-cell biology at CZI. “It exceeds the expectations we set when CZI embarked on funding single-cell biology research and tool development. The technological advances that they demonstrate as well as the biological insight provided are important milestones for the field. These studies are a significant step toward large robust reference resources such as those being pursued by the Human Cell Atlas.”
The Tabula Sapiens project brought together contributions from a wide range of experts, including surgeons and tissue specialists for each organ that was included in the study. “This effort really demonstrates how much potential we can unlock when we embrace the idea of team research,” said Tabula Sapiens Consortium member Angela Oliveira Pisco, Ph.D., associate director of Data Science for CZ Biohub’s Quantitative Cell Science team. “We have coordinated a precise, consistent, and comprehensive effort with more than 160 people, and that in itself is a wonderful feat for science.”
Thanks to a close partnership with Donor Network West, a nonprofit organ procurement organization in Northern California, Tabula Sapiens yields one of the broadest views available of healthy cells across the entire body. “The study maps the building blocks of biospecimens procured in a rather unique project, and demonstrates the remarkable value of non-transplanted tissues and organs for preclinical research,” said Donor Network West’s Ahmad Salehi, M.D., Ph.D., director of research.
In just one session, for example, dozens of surgeons, scientists, and recovery coordinators worked through the night to collect the cells of 17 tissues and organs from a single human donor, within an hour of the withdrawal of life support and the procurement of organs for transplantation. This rapid approach offers the opportunity to study differences in cell types without the degradation of data quality that can occur when using frozen tissue. Also, using samples from a single donor simplifies the data analysis, eliminating the need to control for genetic, age-related, and environmental differences among individuals.
“If we read enough RNA fragments, it’s like having a high-resolution telescope that can see in 25,000 dimensions,” said Bob Jones, a senior research engineer in the Department of Bioengineering at Stanford and member of the Tabula Sapiens Consortium. “It’s incredible detail.”
Tabula Sapiens uniquely includes an analysis of the products of alternative splicing, a cellular process through which differing RNA transcripts can arise from a single gene, leading to numerous protein variants. “Because multiple RNA transcripts can come from each of our 25,000 genes, it becomes very difficult to functionally characterize every single gene and what it can code in the ‘wet lab,'” said Tabula Sapiens Consortium member Julia Salzman, Ph.D., associate professor of biomedical data science and of biochemistry at Stanford. “This kind of research cannot be experimentally driven—it has to be data driven.”
In one surprise already unveiled by Tabula Sapiens, said Salzman, researchers found that sets of housekeeping genes—so called because they have been thought to handle basic functions in much the same way in every cell—likely have many more roles across the body than was previously thought. And in the Science paper, the team reports that CD47, a protein implicated both in cancer and in the buildup of dangerous plaques on artery walls, may differ widely in form from cell to cell, a finding that could guide the development of drugs that are more effective or have fewer side effects. The Science paper also reveals that the gut’s microbiome is “patchy” rather than uniform—Tabula Sapiens data shows that distinct microbial populations exist just a few inches apart in the digestive tract.
“We’re already revealing important new biology that we simply would not have had the possibility of knowing about if it weren’t for Tabula Sapiens,” Salzman said. “With this atlas we have a huge opportunity to improve our understanding of the way the human body works.”
Tabula Sapiens is accessible via a free, easy-to-use data portal, which offers links to all components of the multimodal atlas. “We are committed to making our data easily and widely accessible,” said the Biohub’s Pisco. “Empowering scientists with differing expertise to take full advantage of incredible resources such as Tabula Sapiens will accelerate scientific progress.”
An open-access, user-friendly tool called cellxgene (“cell-by-gene”), developed by CZI’s Science Technology group, means that even scientists with no computational training can employ Tabula Sapiens in their work. The tool, built into the Tabula Sapiens portal, is already helping scientists tackle a variety of questions, such as understanding which cell types are most prone to deleterious gene mutations, how immune-cell populations differ in various organs, and identifying tissues vulnerable to attack by the virus that causes COVID-19.
“Our cellxgene tool is helping scientists, like those in the Tabula Sapiens Consortium, answer fundamental questions about human biology in seconds, not years,” said Phil Smoot, head of science and technology and vice president of engineering at CZI. “We’re excited to see how other scientists will take advantage of this platform to further our understanding of human health and disease.”
You may like to read:
Video Of Bloody-Belly Comb Jelly Pooping Could Be World-First Footage
Alpha-Gal Syndrome: How A Tick’s Saliva Can Make You Allergic To Meat